Summary
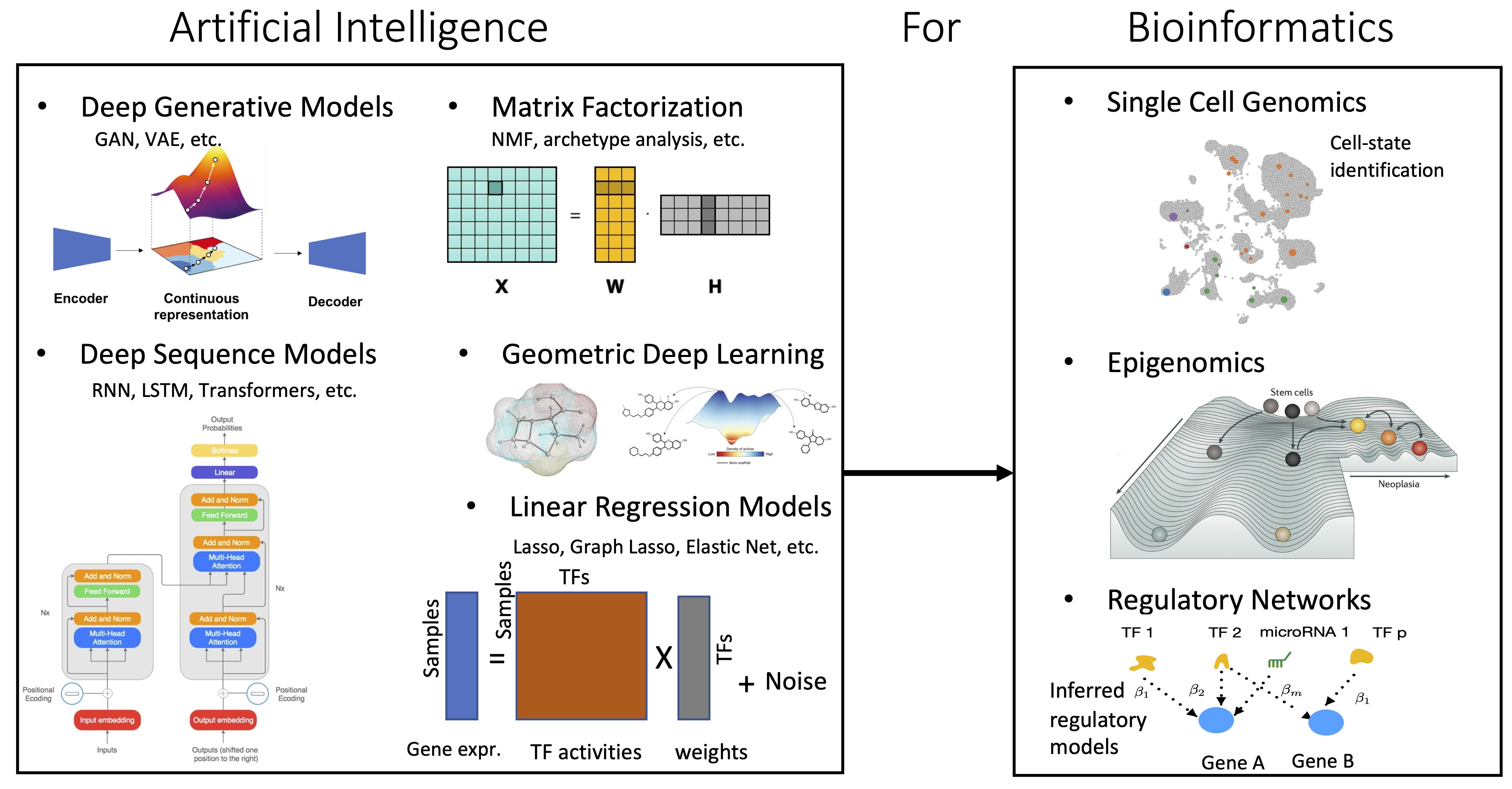
The cell is a highly mysterious intelligent machine that fascinates my imagination and arouses my curiosity. My research revolves around development and application of principled machine learning methods for interpreting complex biological data sets. I love to design novel machine learning algorithms, especially when they allow the analysis of data in ways that have not yet been possible. I am interested in working on fundamental biological problems that would have an impact on our understanding of human cells and disease mechanisms. An ongoing theme in my research is the integration of single cell multi-omics data sets to understand underlying biological processes that contribute to a specific phenotype.
Projects
An efficent regularizaion method to address vanishing gradient in deep neural network archetictures
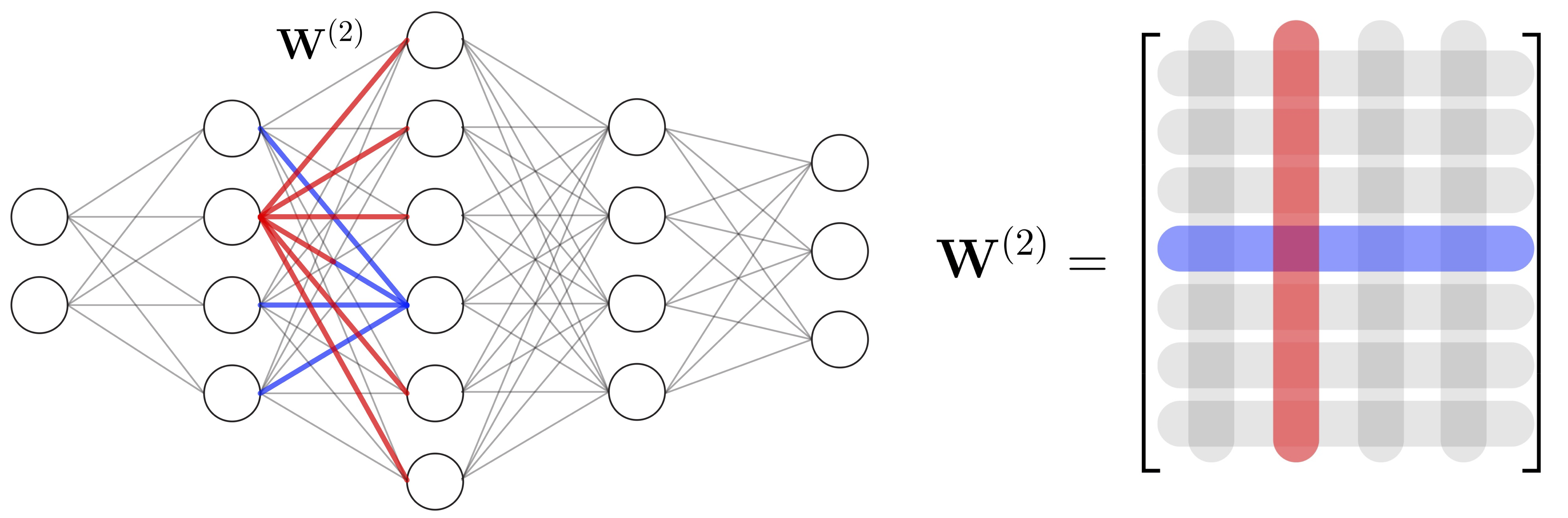
IsoRegNet: A multi-task learning framework to model and analyze transcript isoform regulatory network
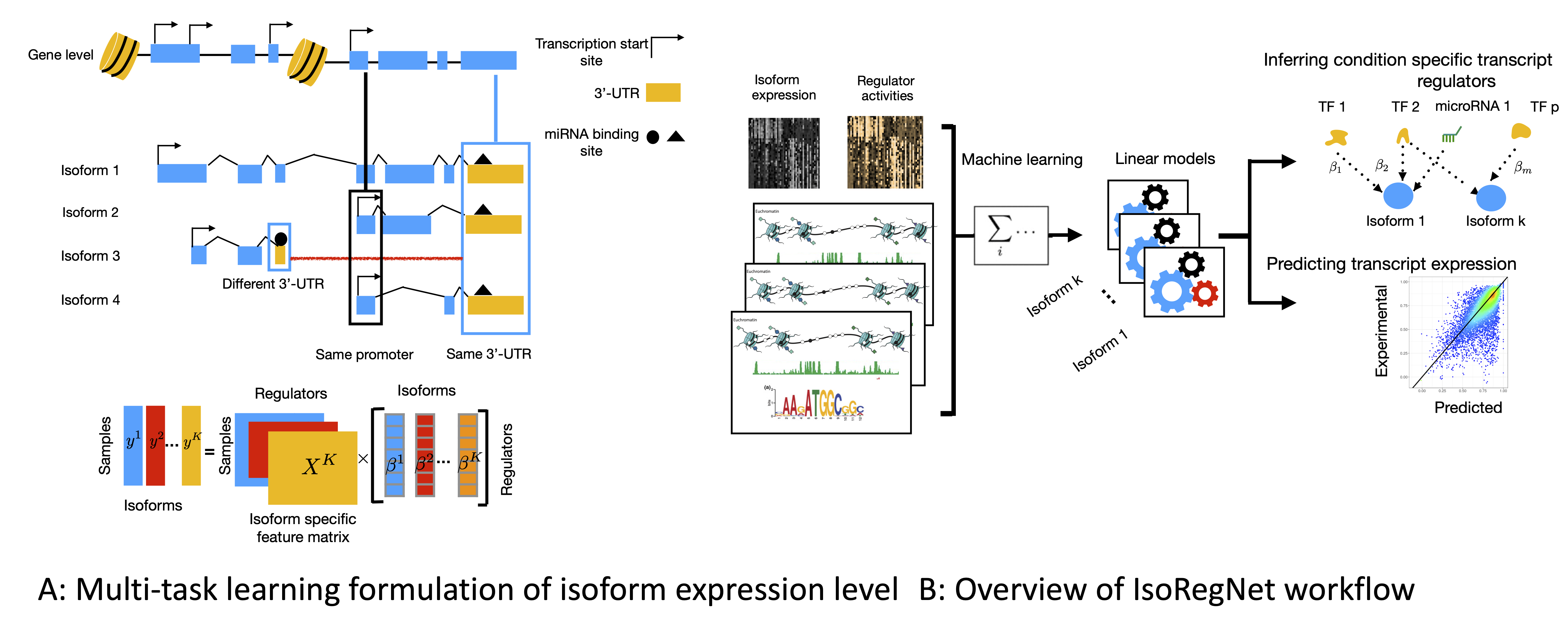
Understanding gene regulation is an important aspect in the context of cell differentiation, disease and environmental changes. Due to the complexity of gene regulation, methods for the inference of gene regulatory networks (GRNs), are often limited to one type of regulator, for example only DNA binding transcription factors or microRNAs. Also the diversity in gene isoforms is rarely taken into account during the construction of GRNs. I've developed a novel principled approach that allows to learn associations between transcriptional and post-transcriptional regulators with genes or their transcript isoforms. Using a novel multi-task formulation we design a regression framework, which allows to overcome some of the noise in isoform expression level estimates, leading to accurate predictions.Inferring genome-wide networks of competing endogenous RNAs
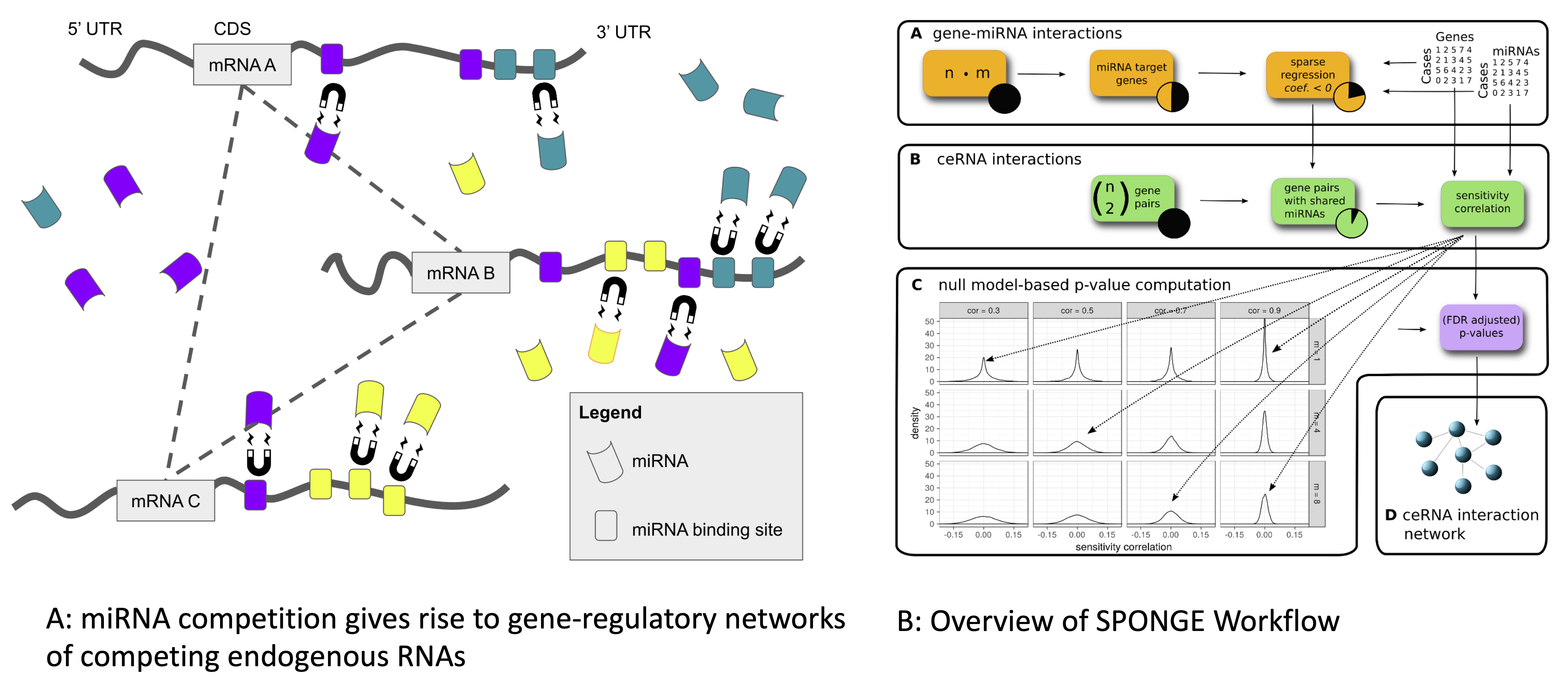 MicroRNAs (miRNAs) are important non-coding post-transcriptional regulators that are involved in many biological processes and human diseases. Individual miRNAs may regulate hundreds of genes, giving rise to a complex gene regulatory network in which transcripts carrying miRNA binding sites act as competing endogenous RNAs (ceRNAs). Several methods for the analysis of ceRNA interactions exist, but these do often not adjust for statistical confounders or address the problem that more than one miRNA interacts with a target transcript. We developed SPONGE, a method for the fast construction of ceRNA networks. SPONGE uses multiple sensitivity correlation, a newly defined measure for which we can estimate a distribution under a null hypothesis. SPONGE can accurately quantify the contribution of multiple miRNAs to a ceRNA interaction with a probabilistic model that addresses previously neglected confounding factors and allows fast P-value calculation, thus outperforming existing approaches. We applied SPONGE to paired miRNA and gene expression data from The Cancer Genome Atlas for studying global effects of miRNA-mediated cross-talk. Our results highlight already established and novel protein-coding and non-coding ceRNAs which could serve as biomarkers in cancer.
MicroRNAs (miRNAs) are important non-coding post-transcriptional regulators that are involved in many biological processes and human diseases. Individual miRNAs may regulate hundreds of genes, giving rise to a complex gene regulatory network in which transcripts carrying miRNA binding sites act as competing endogenous RNAs (ceRNAs). Several methods for the analysis of ceRNA interactions exist, but these do often not adjust for statistical confounders or address the problem that more than one miRNA interacts with a target transcript. We developed SPONGE, a method for the fast construction of ceRNA networks. SPONGE uses multiple sensitivity correlation, a newly defined measure for which we can estimate a distribution under a null hypothesis. SPONGE can accurately quantify the contribution of multiple miRNAs to a ceRNA interaction with a probabilistic model that addresses previously neglected confounding factors and allows fast P-value calculation, thus outperforming existing approaches. We applied SPONGE to paired miRNA and gene expression data from The Cancer Genome Atlas for studying global effects of miRNA-mediated cross-talk. Our results highlight already established and novel protein-coding and non-coding ceRNAs which could serve as biomarkers in cancer.Single cell genomics
Single-cell multi-omics data provides us an opportunity to understand biological processes at the fundamental level. These technologies drive new discoveries and are rapidly advancing, which calls for the constant design and improvement of computational methods for integration, interpretation, and analysis. I develop new methods for single cell data analysis using a combination of rigorous statistical approaches. These methods are necessary to cope with the inherent noise of these technologies and to focus on computationally efficient design and implementation such that resulting method can handle thousands of single cell omics data.
Computational Epigenomics
I would like to continue my work on understanding the system perspective of transcriptional and post-transcriptional gene regulation, answering questions like: What are the rules of enhancer gene targeting? How do structural variations in enhancers effect target gene regulation? Which epigenetic signatures contribute to establishing cell identity? Which regulatory interactions are involved in changing cell identity?
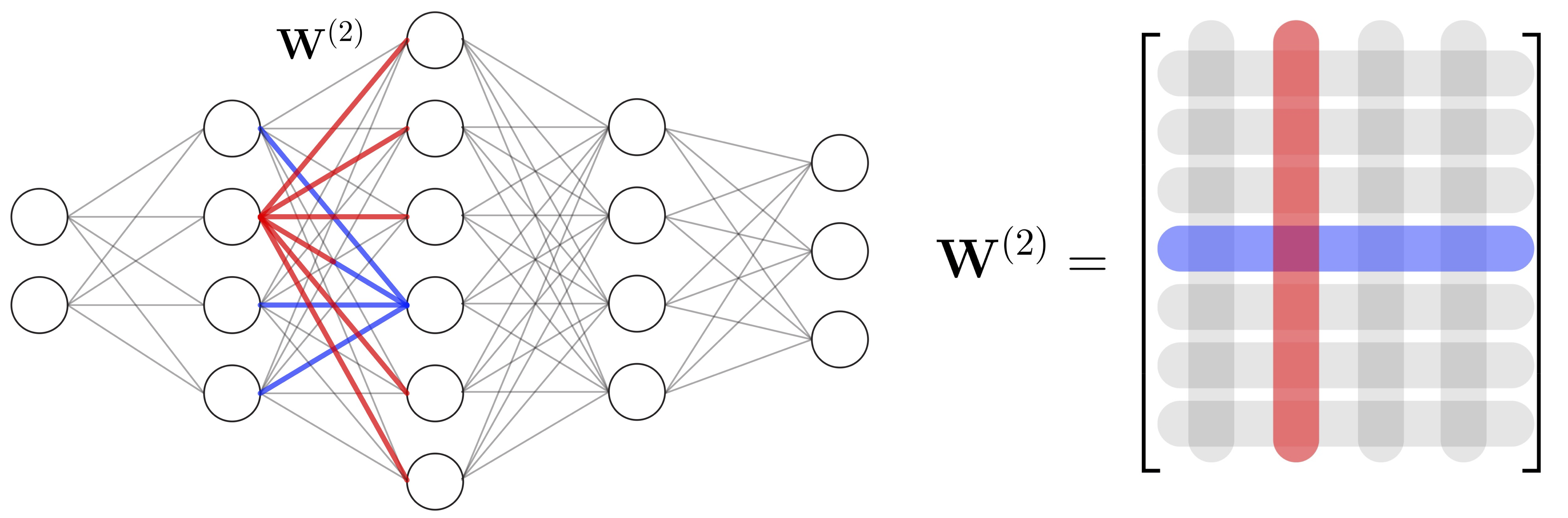
IsoRegNet: A multi-task learning framework to model and analyze transcript isoform regulatory network

Inferring genome-wide networks of competing endogenous RNAs
 MicroRNAs (miRNAs) are important non-coding post-transcriptional regulators that are involved in many biological processes and human diseases. Individual miRNAs may regulate hundreds of genes, giving rise to a complex gene regulatory network in which transcripts carrying miRNA binding sites act as competing endogenous RNAs (ceRNAs). Several methods for the analysis of ceRNA interactions exist, but these do often not adjust for statistical confounders or address the problem that more than one miRNA interacts with a target transcript. We developed SPONGE, a method for the fast construction of ceRNA networks. SPONGE uses multiple sensitivity correlation, a newly defined measure for which we can estimate a distribution under a null hypothesis. SPONGE can accurately quantify the contribution of multiple miRNAs to a ceRNA interaction with a probabilistic model that addresses previously neglected confounding factors and allows fast P-value calculation, thus outperforming existing approaches. We applied SPONGE to paired miRNA and gene expression data from The Cancer Genome Atlas for studying global effects of miRNA-mediated cross-talk. Our results highlight already established and novel protein-coding and non-coding ceRNAs which could serve as biomarkers in cancer.
MicroRNAs (miRNAs) are important non-coding post-transcriptional regulators that are involved in many biological processes and human diseases. Individual miRNAs may regulate hundreds of genes, giving rise to a complex gene regulatory network in which transcripts carrying miRNA binding sites act as competing endogenous RNAs (ceRNAs). Several methods for the analysis of ceRNA interactions exist, but these do often not adjust for statistical confounders or address the problem that more than one miRNA interacts with a target transcript. We developed SPONGE, a method for the fast construction of ceRNA networks. SPONGE uses multiple sensitivity correlation, a newly defined measure for which we can estimate a distribution under a null hypothesis. SPONGE can accurately quantify the contribution of multiple miRNAs to a ceRNA interaction with a probabilistic model that addresses previously neglected confounding factors and allows fast P-value calculation, thus outperforming existing approaches. We applied SPONGE to paired miRNA and gene expression data from The Cancer Genome Atlas for studying global effects of miRNA-mediated cross-talk. Our results highlight already established and novel protein-coding and non-coding ceRNAs which could serve as biomarkers in cancer.Single cell genomics
Single-cell multi-omics data provides us an opportunity to understand biological processes at the fundamental level. These technologies drive new discoveries and are rapidly advancing, which calls for the constant design and improvement of computational methods for integration, interpretation, and analysis. I develop new methods for single cell data analysis using a combination of rigorous statistical approaches. These methods are necessary to cope with the inherent noise of these technologies and to focus on computationally efficient design and implementation such that resulting method can handle thousands of single cell omics data.
Computational Epigenomics
I would like to continue my work on understanding the system perspective of transcriptional and post-transcriptional gene regulation, answering questions like: What are the rules of enhancer gene targeting? How do structural variations in enhancers effect target gene regulation? Which epigenetic signatures contribute to establishing cell identity? Which regulatory interactions are involved in changing cell identity?
